AI accelerates drug discovery from decades to months as Isomorphic Labs’ Drug Design Engine pushes beyond Nobel-Level science
In 2024, Sir Demis Hassabis and John Jumper were awarded the Nobel Prize in Chemistry for AlphaFold, an AI system that solved a fifty-year problem by predicting protein structures at scale.
The achievement felt like a finish line for AI-driven biology. AlphaFold rewired structural biology overnight and reshaped how scientists study life at the molecular level.
Sixteen months later, that milestone already feels like an early chapter.
Key Highlights
-
Isomorphic Labs unveiled IsoDDE, an AI drug design engine built to run discovery directly on a computer, cutting timelines from years to months
-
More than doubles AlphaFold 3 performance on the hardest protein–ligand generalization targets
-
Up to 20× better than Boltz-2 on antibody and biologics benchmarks
-
Outperforms physics-based gold standards on binding affinity prediction, at a fraction of the cost and time
-
Identifies drug binding pockets from sequence alone, including sites that took researchers over 15 years to uncover experimentally
The Isomorphic Labs Drug Design Engine unlocks a new frontier beyond AlphaFold
This week, Isomorphic Labs released a technical report showing that its internal drug design engine has surpassed AlphaFold 3, the latest iteration of the Nobel-winning method. The results point to a sharper shift: AI is no longer stopping at structure prediction. It is a direct step into drug design, with accuracy levels that approach those of experimental methods.
The implication is not abstract. If these systems hold up in real-world programs, the slowest and most expensive phase of medical development may no longer take a decade.
From a Nobel Moment to a Moving Target
AlphaFold changed biology by solving one problem extremely well: predicting the three-dimensional shape of proteins from their amino acid sequences. That alone unlocked years of stalled research. Yet, structure prediction was never the full story behind drug discovery.
Knowing what a protein looks like does not reveal how it binds to a drug, how strong that binding is, where hidden pockets exist, or how the structure shifts when a molecule approaches. Those gaps kept human disease firmly in the hands of lab-heavy workflows.
Isomorphic Labs was formed to close that gap.
Since the public release of AlphaFold 3 in 2024, AI drug discovery has raced ahead. According to Isomorphic Labs’ report, structure alone proved insufficient for real in-silico drug programs. What mattered was whether AI could generalize to unseen chemistry, accurately model binding events, and handle systems that do not behave cleanly.
That is where IsoDDE enters.
What IsoDDE Is and Why “Engine” Matters
IsoDDE is not a single neural network. It is a unified computational engine that integrates multiple predictive tasks into a single system: protein structure prediction, ligand binding, affinity estimation, antibody interactions, and pocket discovery.
The distinction matters. Previous approaches stitched together separate models or relied on physics-based simulations to fill the gaps. Those methods were slow, costly, and fragile when faced with new chemical space.
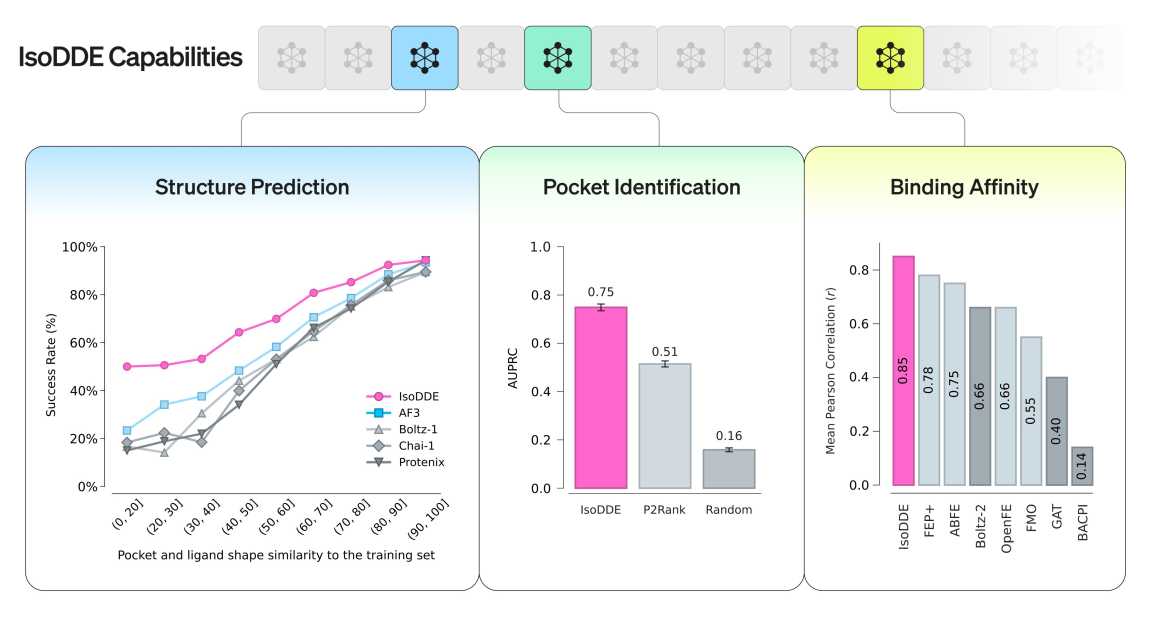
IsoDDE Capabilities (Credit:
IsoDDE runs these predictions in concert, allowing each component to inform the others. That design is what enables the system to handle targets it has never seen before.
In a post on X, Isomorphic Labs said:
“Today we share a technical report demonstrating how our drug design engine achieves a step-change in accuracy for predicting biomolecular structures, more than doubling the performance of AlphaFold 3 on key benchmarks and unlocking rational drug design even for examples it has never seen before.”
The report backs that claim with data.
Doubling AlphaFold 3 on Hard Targets
One of the most striking results appears in protein-ligand generalization tests. These benchmarks are designed to stress models with data outside their training distribution, a known failure point for many AI systems.
IsoDDE more than doubles AlphaFold 3’s accuracy on these tasks, according to the report. That improvement holds even in cases involving induced fits, where proteins change shape as a molecule binds.
Those scenarios have traditionally required long molecular dynamics simulations or direct lab experiments. IsoDDE handles them computationally.
This matters for early drug discovery, where teams often abandon targets because binding behavior is too uncertain to model. Generalization changes that equation.
Finding Drug Pockets From Sequence Alone
Another bottleneck in drug discovery is identifying where drugs bind. Many proteins have pockets that are not obvious from structure alone and only reveal themselves after years of experimental work.
IsoDDE predicts these pockets using the amino acid sequence as its only input. The report shows the engine identifying novel binding sites that were later confirmed experimentally, without prior structural hints.
That capability shifts the search phase from trial and error to targeted exploration. It also expands the range of proteins considered “druggable,” a long-standing constraint in pharmaceutical research.
Antibodies, Biologics, and a New State of the Art
The report also highlights gains in biologics. IsoDDE outperforms existing models on antibody-antigen interface prediction and CDR-H3 loop modeling, two areas where small errors can derail entire programs.
These tasks are critical for therapeutic antibodies, which make up a growing share of modern medicines. Improved accuracy here translates directly into fewer failed candidates downstream.
The authors describe this as a new state of the art in biologics modeling, supported by benchmark comparisons across multiple datasets.
Beating Physics at Binding Affinity
Perhaps the most consequential result appears in binding affinity prediction. Physics-based methods such as free-energy perturbation have long been considered the gold standard for estimating how tightly a drug binds to its target. They are accurate, but painfully slow and expensive.
IsoDDE’s affinity predictions exceed those of physics-based methods in accuracy, according to the report, while running at a fraction of the computational cost.
This is the point at which AI stops acting as a filter and becomes a decision engine. If binding strength can be estimated reliably on a computer, the need to synthesize and test thousands of compounds drops sharply.
This Was Not Built for a Demo
IsoDDE is not new. Isomorphic Labs has been running real drug programs on it for years. The public release offers a window into a system already embedded in active pipelines.
That distinction matters for credibility. The results are not from a single benchmark sprint or isolated dataset. They reflect sustained work across multiple problem classes, validated against established methods.
In an August 2025 interview with 60 Minutes, Hassabis described the stakes clearly:
“So on average, it takes, you know, 10 years and billions of dollars to design just one drug,” he said. “We can maybe reduce that down from years to maybe months or maybe even weeks.”
At the time, the claim sounded ambitious. After IsoDDE, it reads like a roadmap.
What IsoDDE Does, and Does Not Replace
None of this eliminates clinical trials, safety studies, or regulatory review. Human biology remains unpredictable. Drugs still fail for reasons no model can foresee.
What IsoDDE changes is the front end of discovery: the search for viable candidates. That phase consumes the majority of time and capital in drug development. Compressing it reshapes the economics of medicine.
Failures still happen. They just happen faster and earlier.
Why the Nobel Feels Close — and Distant
Hassabis once said most diseases could be cured within ten years. That line drew skepticism, and rightly so. Biology resists timelines.
Yet the distance between that claim and current reality has narrowed. AlphaFold proved AI could read biology. IsoDDE shows AI acting on it.
On X, Hassabis framed the moment carefully:
“The drug design engine we’re building at @IsomorphicLabs is extending the SOTA further across key benchmarks, showing huge progress in accuracy and capabilities critical for in-silico drug discovery. Incredible work from @maxjaderberg and the entire team at Isomorphic Labs!”
The drug design engine we’re building at @IsomorphicLabs is extending the SOTA further across key benchmarks, showing huge progress in accuracy and capabilities critical for in-silico drug discovery. Incredible work from @maxjaderberg and the entire team at Isomorphic Labs! https://t.co/cnnNhjTFtX
— Demis Hassabis (@demishassabis) February 10, 2026
The data support that confidence.
The shelf life of a Nobel Prize has not expired. It has simply been shortened.




