Chinese scientists from Wuhan University just discover a “potentially deadly” new strain of coronavirus called the ‘NeoCoV strain’ with “high fatality and transmission rate”
With billions of people around the world looking to put the pandemic behind them and move on with their lives, a team of Chinese scientists from Wuhan, China just announced they have discovered a “potentially deadly” new strain of coronavirus that they fear could make the jump from animals to humans.
In a research study published on bioRxiv server, the team of researchers from Wuhan University claimed to have “unexpectedly” stumbled upon the new strain, which they’re calling “the NeoCoV strain.” The researchers stated that the strain was originally discovered in South Africa and is a “close relative” of omicron. Although NeoCoV strain “remains enigmatic,” the researchers warned of a “potential bio-safety threat” for humans “with both high fatality and transmission rate.”
“We identified a molecular determinant close to the viral binding interface that 25 restricts human ACE2 from supporting NeoCoV infection, especially around residue Asp338. Conversely, NeoCoV efficiently infects human ACE2 expressing cells after a T510F mutation on the receptor-binding motif (RBM). Notably, the infection could not be cross-neutralized by antibodies targeting SARS-CoV-2 or MERS-CoV. Our study demonstrates the first case of ACE2 usage in MERS-related viruses, shedding light on a potential bio-safety threat of the human emergence of an ACE2 using “MERS-CoV-2” with both high fatality and transmission rate.”
The news of the new NeoCoV strain came shortly after another team of Chinese scientists published new research claiming that the omicron strain may have gestated inside mice. This other team has warned about the “potential bio-safety threat” represented by a new strain of COVID. As part of the study, the researchers said:
“We unexpectedly found that NeoCoV and its close relative, PDF-2180-CoV, can efficiently use some types of bat Angiotensin-converting enzyme 2 (ACE2) and, less favorably, human ACE2 for entry. The two viruses use their spikes’ S1 subunit carboxyl-terminal domains (S1-CTD) for high-affinity and species-specific ACE2 binding.”
It is worth noting this is not the first new strain to emerge since omicron was first discovered by a team in South Africa. On January 8, Cyprus reported a new covid-19 strain called “deltacron.” Unlike previous strains, deltacron is a mutant with attributes of both Delta and Omicron.
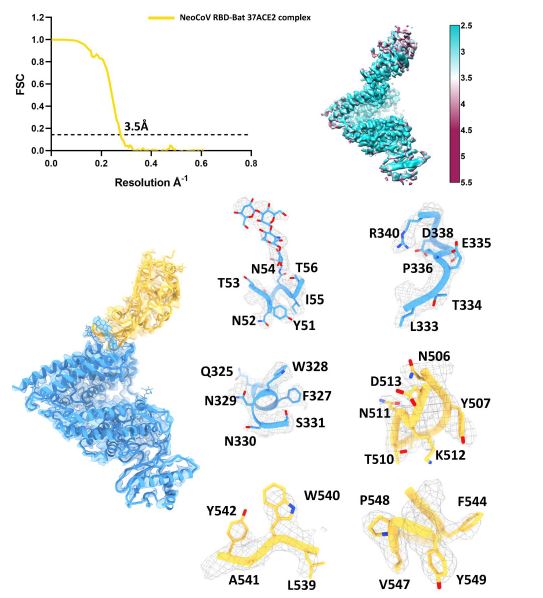
However, cryo-electron microscopy analysis of the new NeoCoV strain revealed a novel coronavirus-ACE2 binding interface and a protein-glycan interaction, distinct from other known ACE2-using viruses,” the researchers said.
In addition, the new strain “can efficiently use some types of bat Angiotensin-converting enzyme 2 (ACE2) and, less favorably, human ACE2 for entry.”
While the strain presently targets bats, the scientists said it has the capability to infect humans as well. And should that happen, it appears the new strain “could not be cross-neutralized by antibodies targeting SARS-CoV-2 or MERS-CoV” meaning natural immunity and vaccine-induced immunity would likely be powerless to stop it.
Below is the Abstract of the study.
Abstract
Middle East Respiratory Syndrome coronavirus (MERS-CoV) and several bat coronaviruses employ Dipeptidyl peptidase-4 (DPP4) as their functional receptors. However, the receptor for NeoCoV, the closest MERS-CoV relative yet discovered in bats, remains enigmatic.
In this study, we unexpectedly found that NeoCoV and its close relative, PDF-2180-CoV, can efficiently use some types of bat Angiotensin-converting enzyme 2 (ACE2) and, less favorably, human ACE2 for entry. The two viruses use their spikes’ S1 subunit carboxyl-terminal domains (S1-CTD) for high-affinity and species-specific ACE2 binding. Cryo-electron microscopy analysis revealed a novel coronavirus-ACE2 binding interface and a protein-glycan interaction, distinct from other known ACE2-using viruses.
We identified a molecular determinant close to the viral binding interface that restricts human ACE2 from supporting NeoCoV infection, especially around residue Asp338. Conversely, NeoCoV efficiently infects human ACE2 expressing cells after a T510F mutation on the receptor-binding motif (RBM). Notably, the infection could not be cross-neutralized by antibodies targeting SARS-CoV-2 or MERS-CoV. Our study demonstrates the first case of ACE2 usage in MERS-related viruses, shedding light on a potential bio-safety threat of the human emergence of an ACE2 using ‘MERS-CoV-2’ with both high fatality and transmission rate.
Competing Interest Statement
The authors have declared no competing interest.
2022.01.24.477490v1.full